Simulation
To apply the theoretical models for brain tissue behavior to relevant clinical problems, we solve the governing equations numerically using the open source finite element library deal.ii. Firstly, we use the computational models to carefully calibrate the model parameters through an inverse parameter identification scheme. This is especially important as, for extremely soft materials such as brain tissue, it is virtually impossible to achieve a homogeneous deformation state during mechanical loading. Once calibrated and validated, the computational tools can be used to tackle scientific issues on the borderline between medicine, biology and mechanics:
The role of mechanics during brain development
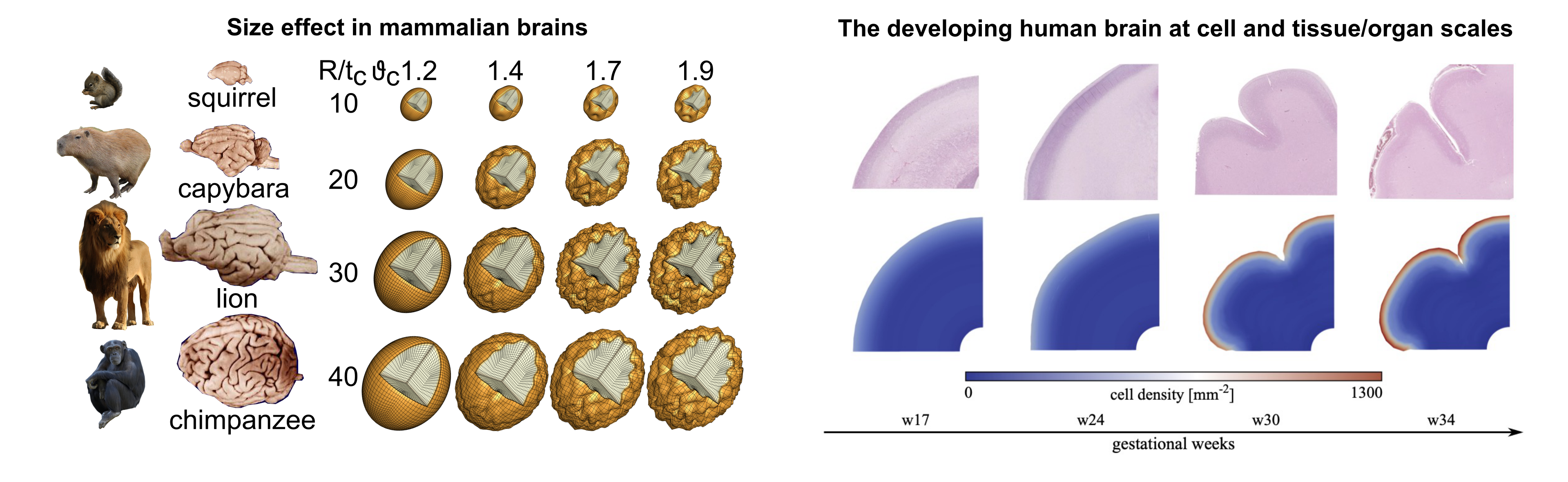
The characteristically folded surface morphology is a classical hallmark of the mammalian brain, as illustrated in Figure 1. During development, the initially smooth surface evolves into an elaborately convoluted pattern, which closely correlates with brain function and serves as a clinical indicator for physiological and pathological conditions. Recent evidence confirms that physical forces play an important role in pattern selection. Studying the correlation between microscopic biological processes occurring within the brain during embryonic life and the macroscopic forces generated is essential for understanding the mechanism behind both normal and abnormal cortical folding. Using our computational model, we investigate essential determinants of brain folding, whether they are of geometrical, mechanical, or biological nature, encompassing cortical thickness, brain geometry, local tissue stiffness, cell division, and cell migration (see Figure 1).
Combining physics and biology holds promise to further advance early diagnostics of cortical malformations and to improve treatment of neurodevelopmental disorders such as epilepsy, autism, and schizophrenia.
Full scale brain simulations
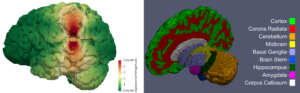
To use the findings from our experimental and modeling investigations in real world applications, we perform simulations using full scale 3-dimensional brain models, as illustrated in Figure 2. These models are created from subject-specific MRI images using the FreeSurfer software suite and an inhouse python code. The resulting models capture the main sulci and gyri of the brain and can be segmented into various regions with distinct mechanical properties.
An important focus of our work is to investigate how different regions with different material properties affect the response of the model under various loading types. We have performed simulations of various surgical procedures (such as brain retraction) and biological processes (such as cerebral atrophy and tumor growth).
By combining our experimental understanding at the tissue level and applying it to a full scale brain model, we hope to contribute to improving the development of protective gear, and to assist the planning of brain surgeries as well as diagnosis and treatment of neurological disorders.
3D Bioprinting
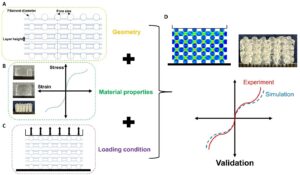
Computational simulations can help predict and tune the mechanical properties of 3D bioprinted constructs – even before fabrication – to save valuable time and cost, as summarized in Figure 3. Like this, a better and faster match between the properties of the biofabricated construct and the target tissue can be achieved. We use the finite element method (FEM) to predict the mechanical properties of different hydrogel mesostructures fabricated through various print patterns and validate our results based on corresponding large-strain experiments under multiple loading modes. This methodology can help save time, material, and cost for future biofabrication applications.
